Merve Haksever, M.Sc.
Department of Scientific Computing
September 2019
Supervisor: Ömür Uğur (Institute of Applied Mathematics, Middle East Technical University, Ankara)
Co-Supervisor: Derya Altıntan (Department of Mathematics, Selçuk University, Konya)
Abstract
Deterministic modeling approach is the traditional way of analyzing the dynamical behavior of a reaction network. However, this approach ignores the discrete and stochastic nature of biochemical processes. In this study, modeling approaches, stochastic simulation algorithms and their relationships to each other are investigated. Then, stochastic and deterministic modeling approaches are applied to biological systems, Lotka-Volterra prey-predator model, Michaelis-Menten enzyme kinetics and JACK-STAT signaling pathway. Also, numerical solutions for ODE system and realizations obtained through stochastic simulation algorithms are compared.
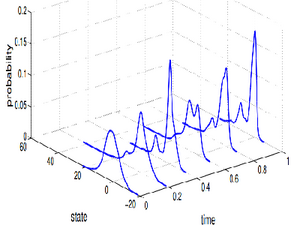
In general, it is not possible to assess all elements of the state vector of biochemical systems. Hence, some statistical models are used to obtain the best estimation. Filtering and smoothing distributions can be obtained via Bayes' rule. However, as an alternative to approximate these distributions Monte Carlo methods might be used. In the second part, bootstrap particle filter algorithm is derived and applied to birth-death process. Estimated probability distribution functions are compared according to number of particles used.
Keywords: mathematical modeling, simulation algorithms, filtering, smoothing